A Spatio-Temporal Link Prediction Pipeline using GC-LSTM for Dynamic Graphs in PyTorch
Background
Dynamic graphs arise in domains where relationships between entities change over time, requiring models that can capture both structural information and temporal evolution. Traditional static graph methods fall short in these settings, motivating the use of spatio-temporal neural architectures. Protein–protein interaction networks, such as those in the DDPIN dataset, exhibit particularly complex and time-varying connectivity patterns, making them a challenging but valuable testbed for link prediction research. Our work explores how gated spatio-temporal models can better forecast future connections in such evolving networks.
Methodology
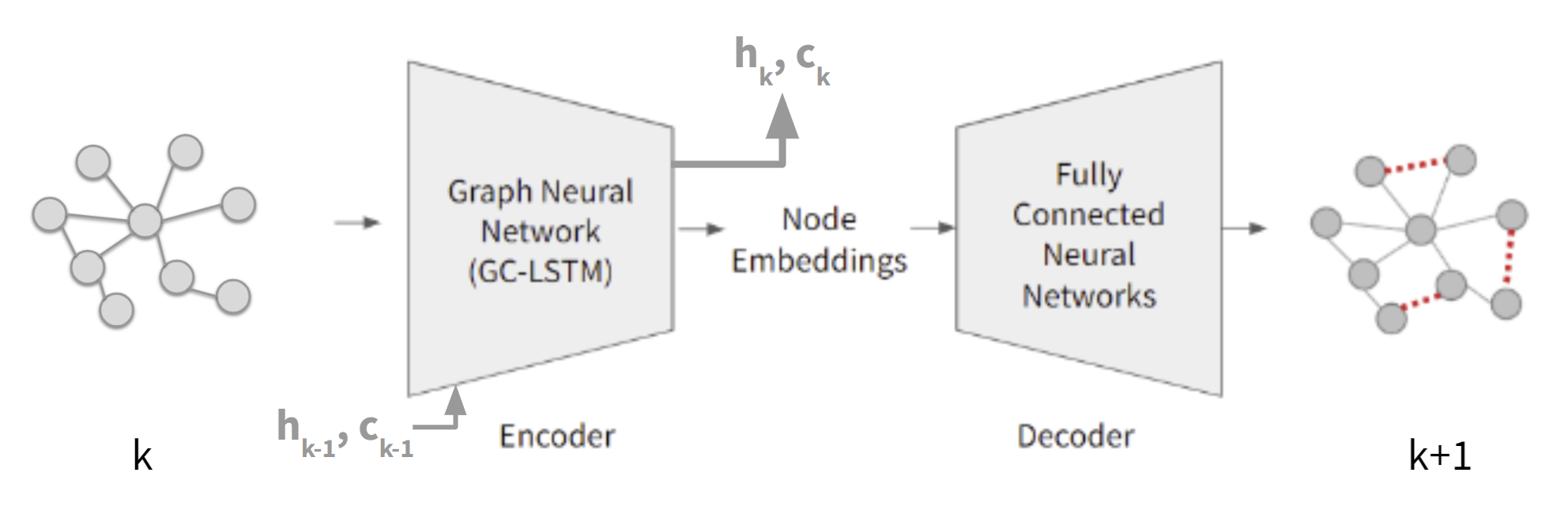
We developed a spatio-temporal link prediction pipeline centered on the GC-LSTM architecture, implemented in PyTorch. The model integrates graph convolutional operations to extract structural features at each time step and LSTM components to capture temporal dependencies across graph snapshots. Using sequences of protein–protein interaction networks from the DDPIN dataset, we trained the model to predict future links by learning from historical connectivity patterns. The pipeline included data preprocessing, temporal batching, negative sampling for training stability, and evaluation metrics tailored to sparse biological graphs. This design allowed the system to effectively learn both localized graph structure and longer-term temporal dynamics.
Findings
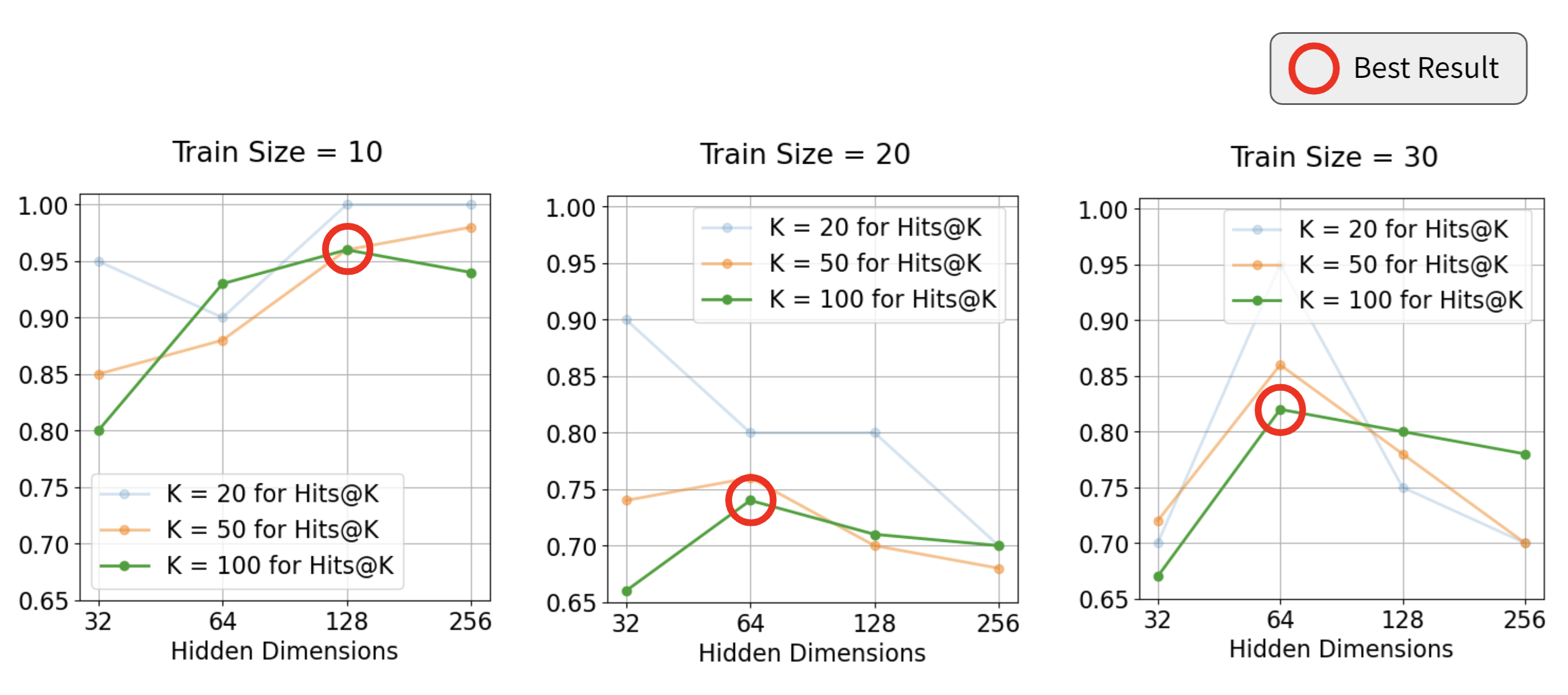
Applied to dynamic protein–protein interaction graphs, the GC-LSTM pipeline achieved over 75% Hits@100 accuracy, indicating strong predictive performance in a biologically complex and highly dynamic domain. The model successfully identified likely future interactions by leveraging both temporal trends and evolving graph structure. Beyond this specific application, our evaluation suggests that the framework generalizes well to other dynamic graph tasks—such as recommendation systems, social-network evolution modeling, and knowledge-graph completion—highlighting its versatility as a tool for forecasting future connections in evolving networks.